REAL-TIME QUANTITATIVE POLYMERASE CHAIN REACTION (Q-PCR)
Attila Szanto
University of Debrecen, Medical and Health Science Center, Department of Biochemistry and Molecular Biology, Life Science Building, Room 3-211, Egyetem ter 1, Debrecen, H-4010, Hungary, Tel.: +36 52 416432, Fax: +36 52 314989
INTRODUCTION TO THE PROCEDURE
Real-time quantitative polymerase chain reaction (Q-PCR) is the most sensitive method for detection and quantification of RNA or DNA molecules. It is a complex method that requires expertise in PCR assay development and optimization. Nevertheless, it is a powerful method with high specificity, sensitivity and reproducibility that can be used in high-throughput studies. Basically, Q-PCR is a fluorescence-based kinetic study where fluorescent signal produced by the proceeding amplification of a specific template is determined in every PCR cycle. The real time determination of the fluorescent signal makes it possible to use the log phase of the PCR for quantification, when the signal correlates with the initial number of template molecules, and at the same time provides high sensitivity when comparing small differences. Another advantage of the technique is that molecules with low-abundancy can also be quantified precisely, allowing the analysis of limiting amounts of cells or tissues.
Reverse transcription
For the analysis of the transcriptome we need quantification of the expressed RNA molecules. There are five commonly used methods for this: Northern-blotting, RNase protection assays, in situ hybridization, microarrays and reverse transcription (RT)-PCR. Among these real time RT-Q-PCR is the most sensitive and suitable method for quantification. To analyze RNA molecules with Q-PCR, they need to be transcribed into cDNA molecules by reverse transcriptase. This can be primed by different methods: gene-specific primers, random primers or oligo-dT primers.
Quantification of cDNA/DNA molecules
Quantitative information is generated from fluorescent signal that correlates with the initial copy number of cDNA or DNA molecules. This signal can be generated from non-specific binding of a fluorescent dye to the amplified nucleic acid (e.g. SYBR Green I) or from sequence specific, fluorescently labeled oligonucleotides, also called probes (e.g. Taqman probes). Both methods can quantify the mRNA/DNA copy numbers of the gene-of-interest, either in an absolute manner when using standard curves (serial dilution of an amplicon with know copy numbers), or in a relative manner when comparing to a specific sample. In both cases normalization to A) a consitutively expressed and not regulated mRNA or B) a gene with stable genomic copy number is essential.
OUTLINE OF THE EXPERIMENTS
In these experiments we will analyze the effects of DMSO priming and 9-cis retinoic acid treatment of CDM1 cells on different levels. Previous experiments are analyzed with one common method, Q-PCR in the following manner:
- Practical
8 – Quantification of ChIP DNA
Epigenetic modifications are analyzed from the chromatin immunoprecipitation experiment (Chapter III, Practical 6).
Each group will analyze their four samples (Input, No-antibody control – NAB, Histone 4 polyacetylation – H4Ac and Histone 3 Lysine 4 Methylation – H3K4Met) by measuring four DNA fragments (transglutaminase 2 core promoter, transglutaminase 2 HR1 enhancer region, CD38 Retinoic Acid Receptor responsive element and 36B4 gene).
- Practical
9 – Quantification of mRNA
Gene expression changes are analyzed by measuring retinoic acid induced target gene expression in the same RNA samples used in the microarray experiment (Chapter I, Practical 1).
Each group will analyze their sample by measuring the expression of two genes-of-interest (both are retinoid receptor target genes: transglutaminase 2 and CD38) and two normalizer genes.
- Practical
10 – Quantification of miRNA
Expression of miRNAs are also analyzed in RNA samples derived from the same cells (Chapter I, Practical 2).
Each group will analyze their sample by measuring the abundance of two miRNAs and one normalizer.
REAL-TIME QUANTITATIVE PCR ANALYSIS - PRACTICALS
Practical 8 – Quantification of ChIP DNA
Each group will analyze four samples:
Input
No Antibody Control (NAB)
H4Ac
H3K4Met
Four genes will be measured from every sample:
Transglutaminase 2 core promoter (Core)
Transglutaminase enhancer HR1 (HR1)
CD38 Retinoic Acid Receptor Response Element (CD38)
36B4 (intron-less gene for control)
Q-PCR analysis will be performed in 384 well plates with a pipetting robot.
Practical 8A: Real-time PCR with TaqMan probes
Checklist:
Samples in strips: Input
NAB
H4Ac
H3K4Met
2x Taqman Master Mix
Primers and Probes
- Thaw the strips with DNA samples from ChIP experiment (Practical 3).
- Mix gently and place on ice.
DNA samples will be measured in triplicates. Six groups will use one plate. Prepare DNA samples and PCR Master mixes according to Figure 1. Note, that six groups prepare four PCR Master mixes.
- Mix the following components for 100 reactions in 1.5 ml Eppendorf tubes for four genes (Core, HR1, CD38 and 36B4, respectively) on ice.
(For one reaction) For 100 reactions
2x Taqman Master Mix 5 ml 500 ml
Forward primer (100 mM) 0.0375 ml 3.75 ml
Reverse primer (100 mM) 0.0375 ml 3.75 ml
Probe (20 mM) 0.0625 ml 6.25 ml
Final Volume (approximately) 5 ml 500 ml
- Mix
- Pipette 60 ml PCR Master mix into each well of an 8-well strip and place it as indicated in Figure 1.

When both the samples and the PCR master mixes are located in the correct position the robot will be started.
When the plate is finished cover it with adhesive optical cover and spin it in the plate centrifuge. Then load plate into the Q-PCR (AB 7900) machine or leave it on ice till starting the PCR.
PCR cycle: 1. 95oC 10 min
2. 95oC 15 sec
3. 60oC 60 sec
Repeat Steps 2 and 3 40 times
- Analyze Q-PCR results with SDS software.
- Export Results Table.
- Analyze data in Excel.
Practical 9 – Quantification of mRNA
Practical 9A: Establishing Standard Curves
Checklist:
Transglutaminase amplicon (6e7)
Marked Eppendorf tubes
Yeast tRNA
2x Taqman Master Mix
Primers and Probe
Nuclease-free water
1. Make a serial dilution of Transglutaminase 2 amplicon.
- Thaw Yeast Transfer RNA (tRNA) Solution (100 ng/ml).
- Take 7 marked Eppendorf tubes:
6e7
6e6
6e5
6e4
6e3
6e2
6e1
- Pipette 18 ml tRNA into each tubes.
- Thaw Tg amplicon (6e7).
- Vortex for 30 sec
- Spin down in Eppendorf centrifuge (15 sec, maximum speed, room temperature)
- Pipette 2 ml of Tg amplicon to the 6e6 tube to make a 10-fold dilution.
- Vortex for 30 sec
- Spin in Eppendorf centrifuge (15 sec, maximum speed)
Repeat these steps with the other tubes (6e5 to 6e1), pipetting Tg amplicon from the dilution made in the previous step to obtain all the 7 log dilution series of the amplicon.
2. Prepare PCR Master mix for Transglutaminase for 30 reactions:
- Mix the following components in a 1.5 ml Eppendorf tube on ice:
(For one reaction) For 30 reactions
2x Taqman Master Mix 10 ml 300 ml
Forward primer (100 mM) 0.075 ml 2.25 ml
Reverse primer (100 mM) 0.075 ml 2.25 ml
Probe (20 mM) 0.125 ml 3.75 ml
Nuclease-free water 7.725 ml 231.75 ml
Final Volume 18 ml
- Mix by vortexing
- Spin down briefly
3. Fill up the 96-well PCR plate:
Samples will be measured in triplicates. Similarly, No Template Control (NTC) will be measured in triplicates as indicated in Figure 2. Note that 4 groups will use one plate.
- Pipette 18 ml PCR Master mix into the indicated wells of a 96 well plate.
- Pipette 2 ml nuclease-free water into the first 3 wells (i.e. A1-3) and continue with the diluted amplicon samples as indicated in Figure 2 (i.e. 6e1 dilution into B1-3 wells).

- When the plate is finished cover it with adhesive optical cover and spin it in plate centrifuge then load plate into the Q-PCR (AB 7900) machine or leave it on ice till starting the PCR.
PCR cycle: 1. 95oC 10 min
2. 95oC 12 s
3. 60oC 30 s
Repeat Steps 2 and 3 40 times
- Analyze Q-PCR results with SDS software.
Practical 9B: Reverse Transcription
The same RNA used for the microarray experiments will be analyzed in this experiment. Note that a portion of the RNA prepared with the Qiagen RNeasy kit was DNase-treated prior to this experiment.
Checklist:
RNA sample from Practical 1
5x First Strand Buffer
dNTP (2.5 mM)
DTT (0.1 M)
Nuclease-free water
Random primer
Reverse Transcriptase will be provided by the assistance
- Thaw RNA samples (Qiagen preps).
- Mix and place on ice.
- Prepare two Master mixes in thin walled 0.2 ml PCR tubes on ice:
For one reaction
RT No-RT
5x First Strand Buffer 8 ml 3 ml
dNTP (2.5 mM) 8 ml 3 ml
DTT (0.1 M) 4 ml 1.5 ml
Random Primer (1 mg/ml) 1.2 ml 0.45 ml
Reverse Transcriptase (200 U/ml) 0.4 ml -
Final Volume 21.6 ml 7.95 ml
Add DNase-treated total RNA (150 ng/ml) 18.4 ml 6.9 ml
Nuclease-free water - 0.15 ml
Final Volume 40 ml 15 ml
- Put tubes into PCR machine
PCR cycle: 25oC 10 min
42oC 2 h
72oC 5 min
4oC hold
cDNA samples can be stored at -20oC.
Practical 9C: Real-time PCR with TaqMan probes
Every group will measure four genes: Cyclophylin
36B4
Transglutaminase 2 (Tgm2)
CD38
Checklist:
cDNA samples (RT and No-RT) from Practical 9B
2x TaqMan Master Mix
Nuclease-free water
Forward, reverse primers and Probes
- Thaw cDNA samples (RT and No-RT).
- Dilute samples 5-fold with Nuclease-free water
- Mix
- Prepare PCR Master mixes for five reactions (one for each gene being measured)
· Mix the following components in a 1.5 ml Eppendorf tube, on ice:
(For one reaction) For 5 reactions
2x Taqman Master Mix 10 ml 50 ml
Forward primer (10 mM) 0.75 ml 3.75 ml
Reverse primer (10 mM) 0.75 ml 3.75 ml
Probe (10 mM) 0.25 ml 1.25 ml
Nuclease-free water 3.25 ml 16.25 ml
Final Volume 15 ml
· Mix by vortexing, then spin down briefly in centrifuge
2. Fill up the 96-well PCR plate:
RT samples will be measured in triplicates while No-RT control will be measured in a single well. Six groups will use one plate.
· Pipette 15 ml PCR Master mix into 4 wells of a 96-well plate as indicated in Figure 3.
· Pipette 5 ml of cDNA (RT) into the first 3 wells and 5 ml sample (No-RT) into the 4th well.
· Repeat these with the other genes as indicated in Figure 3.
· When the plate is finished cover it with adhesive optical cover and spin it in plate centrifuge then load plate into the Q-PCR (AB 7900) machine or leave it on ice till starting the PCR.
PCR cycle: 1. 95oC 10 min
2. 95oC 12 sec
3. 60oC 30 sec
Repeat Steps 2 and 3 40 times
- Analyze Q-PCR results with SDS software.
- Export Results Table.
- Analyze data in Excel.

Alternative protocol for the PCR, if not using a commercial master mix:
For one reaction
10x PCR Buffer 2 ml
MgCl2 (25 mM) 2.4 ml
dNTP (2.5 mM) 1 ml
Forward primer (100 mM) 0.75 ml
Reverse primer (100 mM) 0.75 ml
Probe (20 mM) 0.125 ml
Taq polymerase (5 U/ml) 0.125 ml
ROX reference (50x) 0.4 ml
Nuclease-free water 7.45 ml
Final Volume 15 ml
cDNA 5 ml
Final Volume 20 ml
Practical 10 – Quantification of miRNA
Practical 10A: Reverse Transcription
We are using High Capacity cDNA Archive Kit (Applied Biosystems) to transcribe miRNAs from Trizol-purified total RNA samples. Note that Qiagen preps cannot be used for miRNA analysis because small RNA molecules are lost during that procedure while they are kept during Trizol purification.
The following miRNAs will be measured: hsa-miR-34a
hsa-let-7i
hsa-miR-338
hsa-let-7a
RNU43 (U43 small nucleolar RNA)
Each group will measure two miRNAs and one normalizer small nucleolar RNA:
Group 1, 4, 7 and 10: hsa-miR-34a
hsa-let-7a
RNU43
Group 2, 5, 8 and 11: hsa-miR-7i
hsa-let-7a
RNU43
Group 3, 6, 9 and 12: hsa-miR-338
hsa-let-7a
RNU43
Checklist:
Trizol-isolated RNA samples from Practical 2
10xRT Buffer
dNTP (10 mM)
Nuclease-free water
Reverse Transcriptase, RNAsin and 5xRT Primer will be provided by the assistance
- Thaw RNA samples (Trizol preps).
- Mix and place on ice.
1. Prepare RT master mix for four reactions in a 1.5 ml Eppendorf tube on ice:
· Mix the following components in a 1.5 ml Eppendorf tube, on ice:
RT 1-3
(For one reaction) For 4 reactions
10xRT Buffer 1.2 ml 4.8 ml
dNTP (10 mM) 1.2 ml 4.8 ml
Reverse Transcriptase (50 U/ml) 0.8 ml 3.2 ml
RNAsin (40 U/ml) 0.15 ml 0.6 ml
Nuclease-free water 4.25 ml 17 ml
Final Volume 7.6 ml 30.4 ml
- Mix and pipette 7.6 ml master mix into 3 PCR tubes: RT 1, 2 and 3.
- Add 2.4 ml of the indicated 5x Reverse primer (1-3) to each:
- Add 2 ml of total RNA (20 ng/ml) to each tube
That will give you a final volume of 12 ml.
- Mix
- Put tubes on ice
2. Prepare the following No-RT master mix for four reactions in a 1.5 ml Eppendorf tube on ice.
· Mix the following components in a 1.5 ml Eppendorf tube, on ice:
No-RT 1-3
(For one reaction) For 4 reactions
10x RT Buffer 1.2 ml 4.8 ml
dNTP (10 mM) 1.2 ml 4.8 ml
RNAsin (40 U/ml) 0.15 ml 0.6 ml
Nuclease-free water 5.05 ml 20.2 ml
Final Volume 7.6 ml 30.4 ml
- Mix and pipette 7.6 ml master mix into 3 PCR tubes: No-RT 1, 2 and 3.
- Add 2.4 ml of the indicated 5x Reverse primer (1-3) to each tube
- Add 2 ml of total RNA (20 ng/ml) to each tube
That will give you a final volume of 12 ml.
- Mix tubes gently
- Put tubes into PCR
PCR cycle: 16oC 30 min
42oC 30 min
85oC 5 min
4oC hold
Tubes can be stored at -20oC.
Practical 10B: Real-time PCR with TaqMan probes
Checklist:
Reverse transcribed miRNA samples from Practical 10A
2x TaqMan Master Mix
Nuclease-free water
10x TaqMan miRNA assays will be provided by the assistance
- Thaw cDNA samples (RT1-3 and No-RT1-3)
- Mix
RT samples will be measured in triplicates while No-RT control will be measured in a single well. 6 groups will use one plate. See Figure 4.
1. Prepare PCR Master mixes 1, 2, 3 (one for each miRNA being measured), respectively.
· Mix the following components in a 1.5 ml Eppendorf tube, on ice:
(For one reaction) For 5 reactions
2x Taqman Master Mix 7.5 ml 37.5 ml
10x Taqman miRNA assay 1.5 ml 7.5 ml
Nuclease-free water 4.5 ml 22.5 ml
Final Volume 13.5 ml 67.5 ml
· Mix
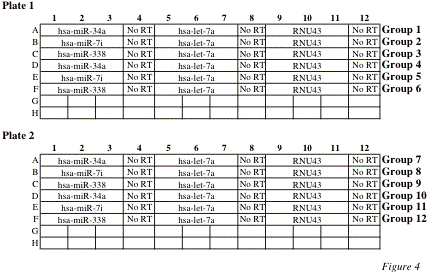
· Pipette 13.5 ml PCR Master mix into 4 wells of a 96 well plate as indicated in Figure 3.
· Pipette 1.5 ml of RT Product (RT) into the first 3 wells and 1.5 ml RT Product (No-RT) into the 4th well as indicated in Figure 4.
· When the plate is finished cover it with adhesive optical cover and spin it in plate centrifuge then load plate into the Q-PCR (AB 7900) machine or leave it on ice till starting the PCR.
PCR cycle: 1. 95oC 10 min
2. 95oC 15 s
3. 60oC 60 s
Repeat Steps 2 and 3 40 times
- Analyze Q-PCR results with SDS software.
- Export Results Table.
- Analyze data in Excel.
Tips and tricks
- When diluting amplicon always use carrier in the solution. Transfer RNA is ideal. At low concentrations the few molecules are easily attached to the surface of the plastic tube when water is used and the probability of degradation is higher.
- When working with concentrated amplicon (>1fmol/ml) use other pipette set than during performing RT or PCR to avoid contamination. Different lab is optimal.
- In vitro transcribed synthetic RNA can be also used as amplicon. In this case radioactive labeling should be used for quantification.
- When many transcripts are measured from the same sample using random primers for RT is more economical than other methods. But if only a few transcripts are measured specific reverse primer can be also used. Always use the same RT method for every gene analyzed. When low quality RNA is analyzed do not use oligo dT.
- While the general amplicon size is below 100 bp the quality of RNA is not critical in Q-PCR.
- In specific reverse primed RT 30 min is enough while in random primed reactions 2h is recommended.
- Use triplicates and No-RT control in Q-PCR measurements.
- Taqman assay is not that expensive and is recommended. Optimization of a SYB Green assay would need several primer group combinations and master mix and could result in a relatively early non-specific signal in the NTCs.
- Do not use Qiagen purified RNA for miRNA quantification.
Reagents
2x SYBR Green Master Mix – Diagenode – GMO-SG2x-A300
2x Taqman Master Mix – Diagenode – GMO-MM2x-A300
Yeast Transfer RNA 25 mg – Invitrogen – 15401-011
Primers – Sigma-Genosys
SuperScript II RNase H-Reverse Transcriptase, 10000U, 200U/ml – Invitrogen – 18064-014
5xFirst Strand Buffer – Invitrogen – Y00146
DTT, 0.1 mM, 500 ml – Invitrogen – Y00147
Random Primer, 300 mg, 3 mg/ml – Invitrogen – 48190-011
20x Reference Dye, 500 ml – Invitrogen – 54881
dATP, 100 mM, 25 mmol – Fermentas – R0141
dCTP, 100 mM, 25 mmol – Fermentas – R0151
dGTP, 100 mM, 25 mmol – Fermentas – R0161
dTTP, 100 mM, 25 mmol – Fermentas – R0171
RiboLock Ribonuclease Inhibitor, 40 U/ml, 2500U – Fermentas – EO0381
Taq DNA Polymerase, 5 U/ml, 500U – Fermentas – EP0402
miRNA Assays – Applied Biosystems
cDNA Archive Kit, 200 rxn, 100 ml – Applied Biosystems – 4322171