NLAB Project foci:
Our laboratory is interested in the epigenomic regulation of cell fate decisions and cellular interactions in physiological processes in health and in pathophysiological processes in various diseases.
The key biological question we are trying to answer is how the identical genome of the various cell types supports the differentiation and function of distinct cell types under different conditions. In particular we are interested in understanding how lipid-regulated transcription factors, among them nuclear receptors, participate in such processes.
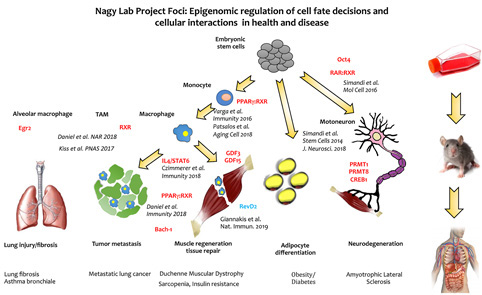
We use cellular models of in vitro differentiation such as monocyte to macrophage differentiation, embryonic stem cell to adipocyte differentiation and embryonic stem cell to neuron differentiation. The overall scheme of the research projects in the combined US-Hungarian laboratory is shown on Figure 1.
VIDEO ABSTRACTS:
--> Giannakis et al Nature Immunology 2019.
--> Czimmerer et al Immunity 2018.
--> Simandi et al Journal of Neuroscience 2018.
--> Daniel et al Immunity 2018.
The following funded projects are carried out in the Debrecen laboratory:
Project 1. Dissecting and aligning the regulatory and effector mechanisms shaping murine M2 macrophages
Macrophages are very heterogeneous and plastic members of the innate immune system. In this project, we are planning to study the molecular background of alternative macrophage polarization-specific functional characteristics using the combination of biochemical, transcriptomic, epigenetic and functional approaches. (1) We will examine the potential regulatory role of lipid sensing nuclear receptors including PPARg, RARa and RXRs in the angiogenic and pro-tumoral activity of alternatively polarized macrophages. (2) We will identify the mechanism and the putative anti-inflammatory role of IL-4/STAT6 signaling-mediated active repression during ex vivo and in vivo alternative macrophage polarization. (3) We are also planing to study the anti-proliferative and/or pro-apoptotic action of IL-4-induced mir-342-3p during the posttranscriptional regulation of macrophage viability.
Project 2. The role of the transcription factor BACH1 in macrophage function and tissue homeostasis
Macrophages are heterogeneous immune cell populations with a remarkable diversity of functions and activation states in different anatomic locations. Their diverse phenotype is the result of a complex interplay between tissue micro-environmental signals and a hardwired differentiation program that determines macrophage identity and fulfills organ-specific and systemic trophic functions. However our knowledge is limited regarding the development of macrophage subtype specification and functional diversity. Our preliminary results pinpoint that the heme-regulated transcription factor, BACH1 is an important regulator in macrophages. It is found to bind extensively to chromatin sites and largely to co-localize with the master regulators PU.1 and CEBP/a. We found that existing BACH1 deficient mouse model likely underestimates BACH1’s function as it is a hypomorph mutant and not a KO.. We propose that BACH1 with its capacity to directly bind metabolites and sense the tissue micro-environment acts as a signal integrator sensing different tissue stimuli and shape transcriptional programs in macrophage subtypes. We will use new genetic tools coupled with transcriptomic and chromatin assays to readdress and dissect the contribution of BACH1 to macrophage specification in vivo. Our proposed experimental plan aims 1) to place BACH1 as part of the core macrophage master regulator complexes regulating macrophage identity/function, 2) to reveal BACH1- dependent homeostatic and inflammatory transcriptional programs in resident and inflammatory macrophages during muscle injury and 3) identify downstream effector molecules and macrophage-tissue crosstalk loops.
Project 3. Dissecting the transcriptional network allowing macrophages to control angiogenesis
The proposed research is the logical continuation of the work described in our publication (Daniel et al 2014). We are going to systematically analyze the interaction of key transcriptional regulators of macrophage polarization and angiogenic activity. We have two specific aims. In aim 1 we will use state of the art epigenomic methods to map the contribution and genome-wide interaction (including 3D), enhancer selection of a set of transcription factors (PPARg, STAT6, EGR2). In the second aim we will study the regulation of two key angiogenic genes VEGFa and HBEGF to understand how macrophage polarization and gene expression lead to an angiogenic phenotype of the cell.